Mind and Matter
Scientific data (in our case e.g. EEG, fMRI or behavioral data) contain patterns that are typically invisible to our eyes. However, sophisticated analysis methods allow us to identify such patterns, and sophisticated graphical methods allow us to visualize them. Analysis results always depend very much on the chosen method, its technical limitations and the method-specific assumptions. Artificial intelligence (AI) methods – and machine learning (ML) in particular – are showing increasing efficiency in the analysis of scientific data. ML methods are now used in neuroscientific research, especially in clinical research, e.g. in the segmentation of 3D tomography images, or in classification tasks in psychopathology and other areas (Milletari et al. 2016, Qureshi et al. 2019, Rad & Furlanello 2016).
However, the often higher performance of ML solutions compared to previous standard methods is accompanied by lengthy training of the artificial network. Powerful computers and, in particular, high-performance graphics cards are becoming increasingly important. In this context, we successfully acquired a Eucor seeding project for the evaluation of clinical data in fall 2019. In addition, we were awarded the contract for a high-performance graphics card (Nvidia Titan V) as part of a tender by NVIDIA. Using this graphics card, we are currently evaluating existing EEG data with AI methods and developing artificial neural networks to analyze the sources on which the EEG is based. Using this graphics card, we are currently analyzing existing EEG data with AI methods (Fig. 1). Using AI, we were able to develop a method for source localization of EEG signals (Hecker et al., 2021, 2023), but also to analyze EEG data more precisely with the help of AI methods (e.g. Wilson et al., 2023).
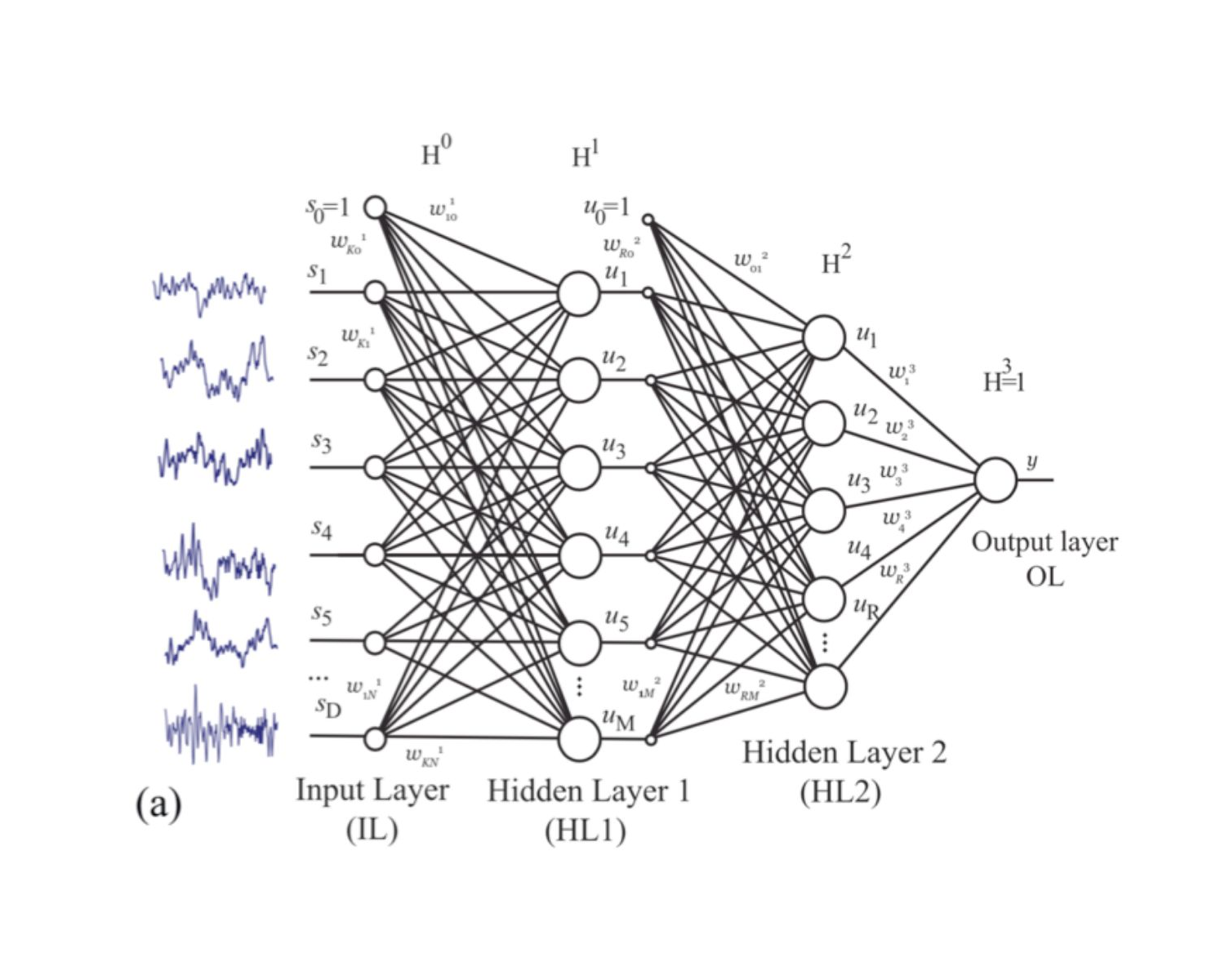
Fig. 1
Example of a feedforward / fully connected neural network (from Hramov et al. 2019). Blue curves (left) are examples of individual EEG curves from a test subject. si, ui and y are neurons of an artificial network. ωi symbolize weights for the information transfer between neurons of successive levels. During the training of such an artificial network, the ωi are gradually changed to improve the network performance.
Publications
Hecker, L., Tebartz Van Elst, L., & Kornmeier, J. (2023). Source localization using recursively applied and projected MUSIC with flexible extent estimation. Frontiers in Neuroscience, 17, 1170862. https://doi.org/10.3389/fnins.2023.1170862
Wilson, M., Hecker, L., Joos, E., Aertsen, A., Tebartz Van Elst, L., & Kornmeier, J. (2023). Spontaneous Necker-cube reversals may not be that spontaneous. Frontiers in Human Neuroscience, 17, 1179081. https://doi.org/10.3389/fnhum.2023.1179081
Hecker, L., Rupprecht, R., Tebartz Van Elst, L., & Kornmeier, J. (2021). ConvDip: A Convolutional Neural Network for Better EEG Source Imaging. Frontiers in Neuroscience, 15, 569918. https://doi.org/10.3389/fnins.2021.569918
Qureshi, M. N. I., Oh, J., & Lee, B. (2019). 3D-CNN based discrimination of schizophrenia using resting-state fMRI. Artificial Intelligence in Medicine, 98, 10-17. https://doi.org/10.1016/j.artmed.2019.06.003
Milletari, F., Navab, N., & Ahmadi, S.-A. (2016). V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation. 2016 Fourth International Conference on 3D Vision (3DV), 565-571. https://doi.org/10.1109/3DV.2016.79
Rad, N. M., & Furlanello, C. (2016). Applying Deep Learning to Stereotypical Motor Movement Detection in Autism Spectrum Disorders. 2016 IEEE 16th International Conference on Data Mining Workshops (ICDMW), 1235-1242. https://doi.org/10.1109/ICDMW.2016.0178